Project 1.8
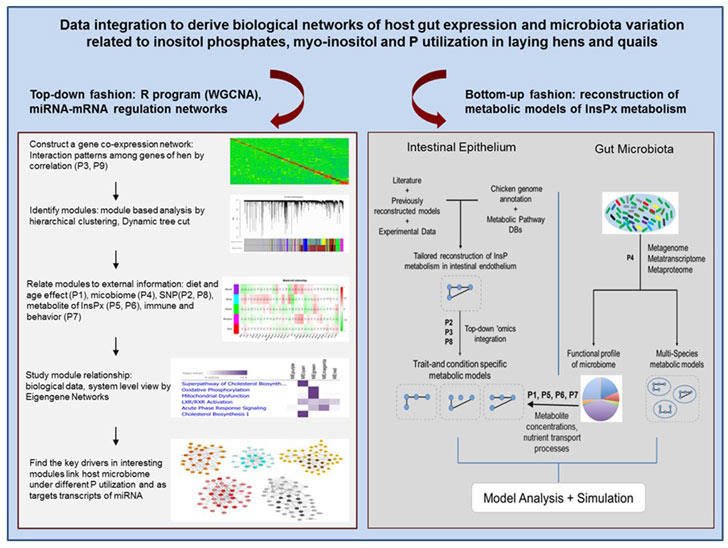
Data integration to derive biological networks of host gut expression and microbiota variation related to inositol phosphates, myo-inositol and P utilization in laying hens and quails
Siriluck Wimmers
There is increasing evidence that the composition of diet and life span play an important role in shaping gut microbiota composition and function which finally has a crucial impact in the development of host physiology and metabolism. Most commonly host-microbiota cross-talk occurs through metabolites of glycolysis/gluconeogenesis, propanoate, starch and sucrose metabolism, and citrate cycle pathways. Also, variable phosphorus (P) supply was shown to impact the microbiota composition.
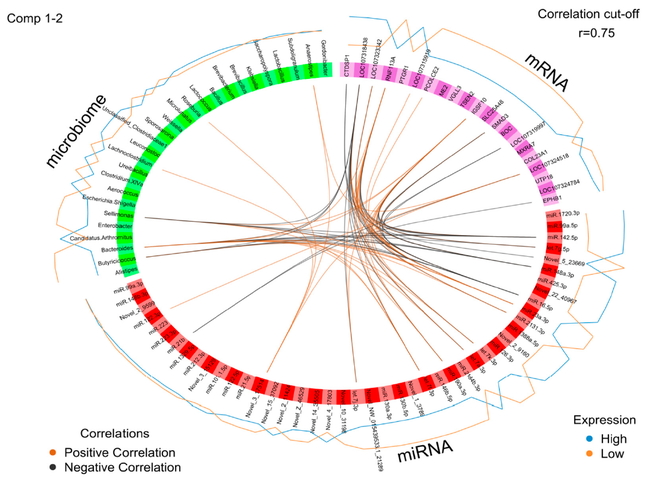
Moreover, recent studies emphasize the role of host genetic background, as both genome and mitochondria can be targeted by microbiota. Host-microbiota relationship can also be mediated by molecules like microRNAs (miRNA) as demonstrated in recent studies. miRNAs are promising candidates for regulatory interactions between the host and the gut microbiota, due to their strong evolutionary conservation.
To draw a more comprehensive view of biological processes, experimental data based on genetic and non-genetic variations in the formation of inositol phosphate derivatives and myo-inositol by endogenous phytases, and their physiological relevance in fowl have to be integrated. Additionally, the important molecules mediating host-microbiome interactions, e.g., miRNAs will be analyzed and integrated with regulatory biological networks under different life span and dietary variables and genetic backgrounds.
This project will enable us to derive biological networks of host expression and microbiota variation related to inositol phosphates, myo-inositol and P utilization in laying hens and quails.