Project 1.7
Mitochondrial haplotypes and phenotypes in laying hens
Martin Hasselmann / Korinna Huber
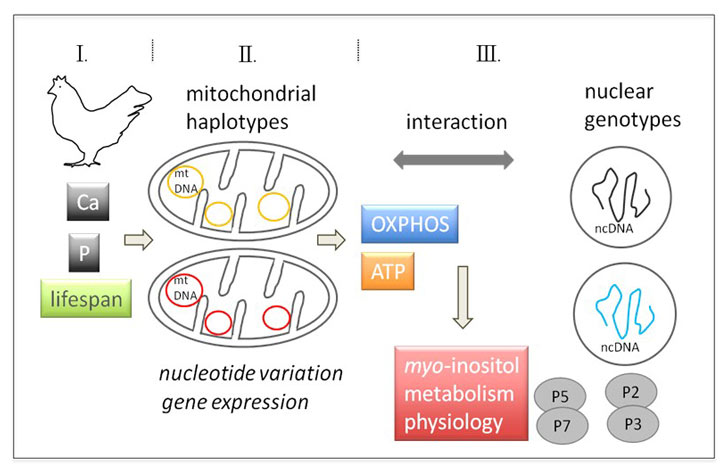
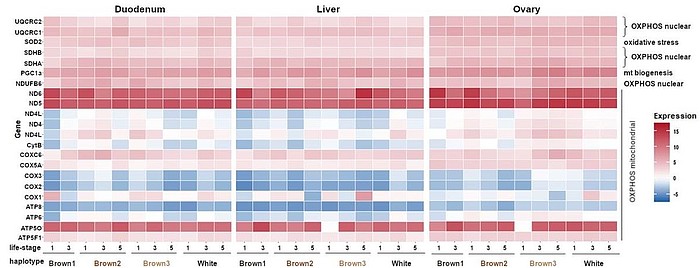
Mitochondria are involved in several organismic key processes; hence the in-depth characterization of mtDNA haplotypes and mt gene expression in hens with efficient phosphorus (P) utilization and myo-inositol metabolism is of utmost importance. Myo-inositol is known to modulate mitochondrial functionality by so far unknown mechanisms related to autophagy and mitophagy.
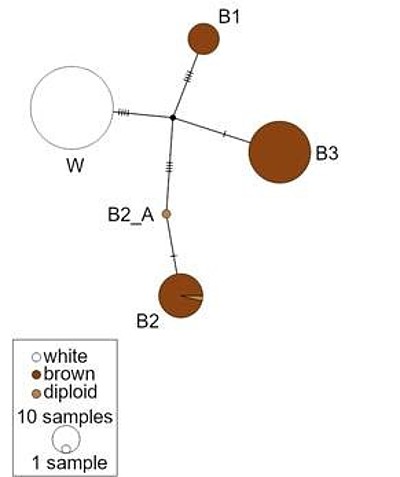
The project will obtain whole mitochondrial genome haplotypes for the two strains of laying hens. This approach determines potential mutations that arose during the genetic selection process in laying hens affecting key enzyme activities. The data will be combined with those of genome analyses obtained in other projects of the Research Unit to find associations of interacting loci depending on the nuclear background.
Further, mtDNA haplotypes are associated with the molecular information regarding metabolomics profiles assessed in another project. Thus, comprehensive insights into the functional importance of mitochondria in respect to P utilization and myo-inositol metabolism in hens will be gained.